 fizer, the maker of the dreaded but successful mRNA vaccine, is
testing a new anti-COVID drug named PF-07321332, popularly dubbed
Pfizermectin. Fact-checkers, lacking any concept of irony,
helpfully jumped in to let us know it's structurally different from
Ivermectin, a drug derided in the press as a “horse dewormer.”
Merck has also developed a drug named Molnupiravir.
fizer, the maker of the dreaded but successful mRNA vaccine, is
testing a new anti-COVID drug named PF-07321332, popularly dubbed
Pfizermectin. Fact-checkers, lacking any concept of irony,
helpfully jumped in to let us know it's structurally different from
Ivermectin, a drug derided in the press as a “horse dewormer.”
Merck has also developed a drug named Molnupiravir.
PF-07321332
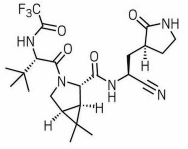
PF-07321332, aka Pfizermectin
PF-07321332 is the most likely drug to succeed. If it works, it'll be our first real treatment for COVID. It's not a reformulated version of ivermectin. Unlike ivermectin, PF-07321332 is not even a macrocyclic, which means the structure doesn't have a large closed loop. The important piece is the C≡N group at the lower right, called an nitrile group. PF-07321332 would probably be administered along with ritonavir to increase the blood levels of PF-07321332.
Proteases
PF-07321332 is an inhibitor of 3-chymotrypsin-like-protease (3CLpro), which is an enzyme encoded by the coronavirus [1]. The viral (+)ssRNA strand is translated into a huge single protein (800 kDa), which must be cleaved by proteases 3CLpro and papain-like protease (PLpro) to create the 16 so-called non-structural proteins, or NSPs, needed for viral replication. Thus, 3CLpro is a better target than TMPRSS2 or furin, which are cellular proteases that cleave the S1 and S2 subunits of the spike protein to allow entry into the cell, because these enzymes serve important functions in the cell.
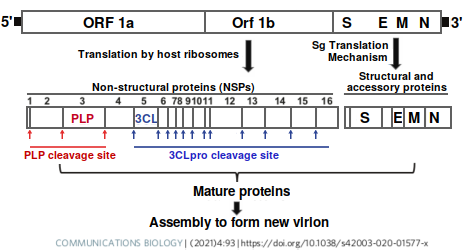
Protease cleavage sites in SARS-CoV-2 (redrawn from [1])
Computer modeling simulations find that many drugs thought to be ineffective, including ivermectin and hydroxychloroquine, are reasonably good inhibitors of 3CLpro, and in vitro measurements confirm it. In the table below, theoretical binding affinity is given by Kcal/mol, so the more negative the number the stronger it binds. Any number lower than −9 is considered promising.
Weird results sometimes show up in computer models. For instance, Atorvastatin, a cholesterol-reducing drug, was also predicted to bind to the protease. Theoretical binding affinity to some place on the protein does not necessarily mean effective inhibition, and Lopinavir / Ritonavir failed in clinical trials. The remarkably high affinity of PF-07321332 is due to the highly reactive nitrile group, which is said to form a covalent bond with the protein [4,5].
| Drug | Kcal/mol | Kd (μM) | IC50 (μM) |
|---|---|---|---|
| Remdesivir | −5.95 | 64.1 | |
| Hydroxychloroquine | −7.04 | 10.9 | |
| Paritaprevir | −7.43 | 5.8 | 73.4 |
| Ivermectin | −7.74 | 3.5 | 21.5 |
| Atorvastatin | −7.99 | 2.3 | |
| Lopinavir | −9.10 | 0.39 | >50 |
| Danoprevir | −9.27 | 0.29 | >50 |
| Ledipasvir | −9.53 | 0.19 | |
| Micafungin | −9.60 | 0.17 | 47.6 |
| Asunaprevir | −9.64 | 0.16 | >50 |
| Ritonavir | −9.95 | 0.10 | >50 |
| PF-07321332 | −24.37 | 6.7×10−12 |
Values for PF are from [2], others are from [3–5]. Ref [2] estimates −8.0 for lopinavir and −10.4 for ritonavir. Relative standard deviations for these terms are on the order of 20%. IC50 values are from pharmacological measurements and show the concentration needed to inhibit the enzyme by 50%.
What conclusion can we draw from this? First, computer simulations don't always match up with what happens in the cell, which is why we still need direct measurements. Second, if people think scientists are ignoring HCQ and ivermectin, they're mistaken.
Molnupiravir
Merck's Molnupiravir (MK-4482) works by inducing mutagenesis. Even the dull Nature Structural & Molecular Biology titled their article “Coding for catastrophe”[6] (gruesome molecular details in [7]). Of course, the authors mean catastrophe for the virus's genome, not ours . . . hopefully.
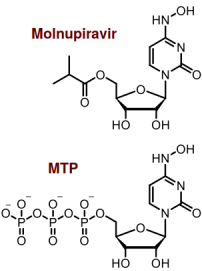
Molnupiravir and MTP. Technically, MTP is β-D-N4-hydroxycytidine (NHC) triphosphate, but (almost) nobody calls it that
Like Remdesivir, Molnupiravir targets RNA-dependent RNA polymerase (RdRp), which is a protein coded by the virus that makes copies of the virus's (+)ssRNA molecule. Structurally, Molnupiravir is a prodrug form of β-D-N4-hydroxycytidine (NHC), which is an analogue of one of the four ribonucleotides that make up the RNA strand. The active form of the drug is loosely called molnupiravir triphosphate (MTP), which is highly charged, so it can't enter the cell. That's why molnupiravir is made with an OH group esterified to isobutyric acid, making it a prodrug. Once molnupiravir enters the cell, the cell chonks off the organic acid and converts the remainder to MTP, which is the active form of the drug. MTP competes with CTP and is incorporated into the new virus RNA.
The clever thing about MTP is that unlike C, which only pairs with G, M tautomerizes (which means it rapidly interconverts into one of two different forms) in such a way as to mimic either C or U. Thus, it can substitute for C or U and pair with either G or A. Since there are 9594 U's and 5492 C's in Sars-CoV-2, this effectively turns the viral RNA into nonsensical RNA goo. Unlike remdesivir, which stalls the polymerase when it gets incorporated, with MTP the enzyme just keeps on going.
According to my calculations, assuming MTP works perfectly, after only two cycles 50% of the bases will be changed (see graph). The percentage doesn't go to 25% (which would be perfectly random since there are four bases in RNA), but approaches a minimum of 37.5 percent, or 1.5×25%.
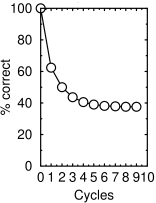
Calculated percentage of correct bases on (+) strand as a function of number of replication cycles in the presence of molnupiravir, assuming it works perfectly as described in ref.7
The drug is 100 times more active than ribavirin, having an IC50 of 0.15μM[11]. There are only a couple of teensy-weensy little problems. One is that the number of mutations is concentration-dependent. So if some cells happened to get only a low dose of MTP, it would turn the cell into a mutant virus factory in which many new, possibly viable, mutant viruses with a handful of mutations apiece were efficiently manufactured. If one of those turned out to be highly pathogenic, it would be very embarrassing.
Another possible concern is whether molnupiravir could get converted to the 2′-deoxyribose form of NHC, thus forming 2′deoxy-MTP. This would cause mutations in the human cells that could lead to cancer and birth defects. Humans have an enzyme called ribonucleotide reductase that does this. Using a cell test, a group at the University of North Carolina found evidence that NHC is indeed genotoxic[12]. If so, it means it would only be useful in desperate life-threatening cases, and the drug would not be approved for general use (though with the FDA approving practically anything that walks in the door these days, emergency approval is probably likely).
In a letter to the editors of J. Infect. Dis., Merck scientists disputed those findings. Merck claimed that in their assay, mutation rates are not different from the rates in control animals. Merck also says the drug has a favorable safety and tolerability profile and that it reduces mild to moderate hospitalizations by 50 percent.
In a strongly-worded rebuttal, Zhou et al. defended their results, pointing out that mutagenesis is not an acute toxicity, but would only be revealed over a long period in cancer rates and germline mutations, so the longer incubation time that they used is needed to see them. They conclude that molnupiravir will lead to mutations in host DNA in dividing cells and that its use poses “unknown long-term risks.”
It should be noted that half the authors, including RS Baric (whose name is familiar from the virus gain-of-function debate), do not appear on the rebuttal. Genotoxic drugs are commonly used to treat cancer, so approval would depend on how the FDA sees the risk/benefit ratio. Nonetheless, the authors have raised serious questions that will need to be resolved by more scientific research.
The third possible problem is what would happen if molnupiravir got loose in the mitochondria. If it did, it would interfere with mitochondrial DNA-dependent RNA polymerase (mtRNAP, aka POLRMT), which is needed to transcribe mitochondrial DNA [8,9] to make ribosomal, messenger, and transfer RNA in the mitochondria. This would screw up mitochondrial function. It could also screw up dinucleotide signaling, which is important in . . . waaaaait for it . . . inflammation [10]. It's always inflammation.
1 Mody, V., Ho, J., Wills, S. et al. Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. Commun Biol 4, 93 (2021). https://doi.org/10.1038/s42003-020-01577-x https://www.nature.com/articles/s42003-020-01577-x
2 Ahmad B, Batool M, Ain QU, Kim MS, Choi S. Exploring the Binding Mechanism of PF-07321332 SARS-CoV-2 Protease Inhibitor through Molecular Dynamics and Binding Free Energy Simulations. Int J Mol Sci. 2021 Aug 24;22(17):9124. doi: 10.3390/ijms22179124. PMID: 34502033; PMCID: PMC8430524.
3 Vandyck K, Deval J. Considerations for the discovery and development of 3-chymotrypsin-like cysteine protease inhibitors targeting SARS-CoV-2 infection. Curr Opin Virol. 2021;49:36–40. doi:10.1016/j.coviro.2021.04.006
4 Pavan M, Bolcato G, Bassani D, Sturlese M, Moro S. Supervised Molecular Dynamics (SuMD) Insights into the mechanism of action of SARS-CoV-2 main protease inhibitor PF-07321332. J Enzyme Inhib Med Chem. 2021 Dec;36(1):1646–1650. doi: 10.1080/14756366.2021.1954919. PMID: 34289752; PMCID: PMC8300928.
5 Ramos-Guzmán CA, Ruiz-Pernía JJ, Tunón I. Computational simulations on the binding and reactivity of a nitrile inhibitor of the SARS-CoV-2 main protease. Chem Commun (Camb). 2021 Sep 9;57(72):9096–9099. doi: 10.1039/d1cc03953a. PMID: 34498651. https://pubs.rsc.org/en/content/articlelanding/2021/CC/D1CC03953A (paywalled, inaccessible)
6 Malone B, Campbell EA. Molnupiravir: coding for catastrophe. Nat Struct Mol Biol. 2021 Sep;28(9):706–708. doi: 10.1038/s41594-021-00657-8. PMID: 34518697.
7 Kabinger F, Stiller C, Schmitzová J, Dienemann C, Kokic G, Hillen HS, Höbartner C, Cramer P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat Struct Mol Biol. 2021 Sep;28(9):740–746. doi: 10.1038/s41594-021-00651-0. PMID: 34381216; PMCID: PMC8437801.
8 Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011 Sep 25;478(7368):269–273. doi: 10.1038/nature10435. PMID: 21947009.
9 Arnold JJ, Smidansky ED, Moustafa IM, Cameron CE. Human mitochondrial RNA polymerase: structure-function, mechanism and inhibition. Biochim Biophys Acta. 2012 Sep–Oct;1819(9–10):948–960. doi: 10.1016/j.bbagrm.2012.04.002. PMID: 22551784.
10 Menzel S, Schwarz N, Haag F, Koch-Nolte F. Nanobody-Based Biologics for Modulating Purinergic Signaling in Inflammation and Immunity. Front Pharmacol. 2018 Mar 27;9:266. doi: 10.3389/fphar.2018.00266. PMID: 29636685; PMCID: PMC5880931.
11 Sheahan TP, Sims AC, Zhou S, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12(541):eabb5883. doi:10.1126/scitranslmed.abb5883
12 Zhou S, Hill CS, Sarkar S, Tse LV, Woodburn BMD, Schinazi RF, Sheahan TP, Baric RS, Heise MT, Swanstrom R. β-d-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells. J Infect Dis. 2021 Aug 2;224(3):415–419. doi: 10.1093/infdis/jiab247. PMID: 33961695; PMCID: PMC8136050.
oct 08 2021, 6:35 am. updated oct 10 and oct 19 2021
