Image forensic testing in imal
This page describes how to analyze scientific images to detect image manipulation using Imal (the Image Measurement and Analysis Laboratory).
This software is free and open-source. It runs in Linux using Motif and compiles easily with g++ in Debian and most other Linux distributions. A pre-compiled dynamically linked version is available.
It is intended for two purposes: to provide evidence to refute allegations of image fraud, and to protect researchers by flagging images before publication that could be misinterpreted as fraudulent by scientific journals or overenthusiastic Internet sleuths. Often, journals rely on overhyped software packages that claim to be able to detect flaws in an image. Unless you take active means to protect yourself, you run the risk that an opaque algorithm could be used to falsely accuse you of a misdeed and leave you with no way to refute the allegation.
Why you need it
I knew an brilliant professor who was highly skilled at his job but had little knowledge of image forensics. He was caught unprepared by an avalanche of false allegations. His colleagues abandoned him for fear of being accused themselves. He struggled to gain the necessary background in an attempt to verify or refute the claims, but he was too late and the university lost an admired administrative leader. The careers of everyone in his department were damaged, including those whose only connection with the professor was to have cited the his work before it came under suspicion.
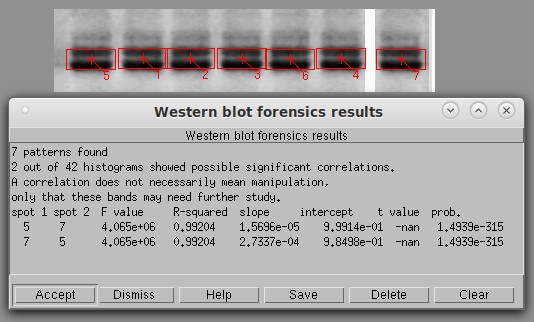
Basic histogram analysis of a typical Western blot. The software automatically identifies the bands and draws red boxes around them. It then calculates the degree of similarity between the bands using a technique that cannot be fooled by vertical or horizontal flipping. The first six lanes on the left might appear to be flipped copies of each other (i.e., 1 a copy of 5, 3 a flipped copy of 2, etc). To test this, I created lane 7 as a positive control by flipping a copy of lane 5 and ran a histogram test with a threshold of 0.90. Only the known positive control (5 and 7) was significant, unequivocally proving that the bands are not copies of each other. The next highest pair was lanes 3 and 6, with an r2 of only 0.689.
Testing a scientific image is serious business. It is essential for every scientist and technician to be familiar with the principles of image forensics. You must also be intimately familiar with the algorithms used by the software and their strengths and weaknesses to have a chance of defending yourself against a malicious allegation. Scientific journals use a variety of unknown and sometimes unvalidated software, as editors are readily swayed by hackers who provide colorful graphs. They are easily fooled by claims that the software uses “artificial intelligence” (while in fact it may actually be using a hundred-year-old numerical algorithm) but readily generates false positives. As a professional who deals with scientific images, you must know enough to be able to defend yourself before an allegation is made. Remember that university bureaucrats will not take your side. By the time the bureaucrats get around to telling you your article is being retracted because of a “suspicious” image, it will be too late to rebut the accusations if you are unprepared and unknowledgeable.
General guidelines
As with any scientific procedure, the goal is not to arrive at the conclusion you want but to find out the truth. If your image should turn out to have been tampered with, you will need solid evidence of what was done before confronting the miscreant. If your image is good, your lawyer will need solid evidence to present to the court in your defamation suit.
This software has been tested against a variety of real and simulated Western blot images. The author is an expert biochemist who has run thousands of Western blots, but there are no guarantees. Feedback and bug reports are appreciated.
This software is not designed for use against microscope images. It may have value on such images, but has been untested.
Here are some guidelines.
Always work from a copy and keep the original write-protected.
Document every step so others can repeat your analysis and get the same result.
Present the results as statistical figures and probabilities wherever possible and emphasize that definitive conclusions can never be obtained from image analysis.
Work with the original 16-bit image if at all possible. Do not attempt to analyze images in JPEG format or other low-quality images.
In each test, you need a positive and negative control just as if you were doing an experiment.
Practice creating falsified images to familiarize yourself with what the software can and cannot do. This will also help you think like a miscreant. Some of them are very skillful and creative, but most simply flip entire rows of Westerns, change their height and width, and—occasionally—adjust the contrast. Often they don't even think to eliminate telltale smudges that give them away.
If an image turns out to have been manipulated by a co-worker, do not destroy it but quarantine it and preserve your documentation as legal evidence.
The three basic techniques
There are three basic ways of analyzing a Western blot: by shape, by pixel values, and by artifact detection. Shape analysis uses some mathematical transform such as PCA (principal component analysis) or wavelet decomposition to obtain a numerical fingerprint of the image and compare it with other images or other parts of the same image. Pixel value analysis examines the relationship among the pixel values, i.e. shades of gray, in an image. Artifact detection looks for telltale signs left by a miscreant. An analysis is not complete unless all three techniques are used. They complement each other: all three are looking for different types of artifacts, and they go about it in different ways.
Image analysis is hypothesis-driven. There is no simple, universal way of comparing images. We might hypothesize, for example, that the band in a Western blot was a (possibly distorted) copy of another blot in the same paper or an earlier one, and use the computer to test the hypothesis. The same algorithm could not test the hypothesis that it was a copy of some blot that we have never seen. To test that, all we can do is look for inconsistencies within the image itself. Few investigators would ever think to test the hypothesis that two blots look similar because they were both dropped on the street and run over by the same 2016 Chevy Impala, but it has probably happened.
General Procedure
Open the image in Imal. Click About→About the Image and note the pixel depth, file format as identified by the computer, color type, and number of grayscale levels in the image. An absolute minimum is 100 grayscale levels. A good image will have several thousand.
Check under Edit→Highlight Saturated Pixels to make sure that your image does not have excessive numbers of saturated pixels. If there are many of these on the important parts of your image, it indicates clipping and could produce an incorrect result.
Crop off any labels or junk around the edge. This could be incorrectly identified as a band and clutter up the results.
If the image is not already 16-bit/pixel grayscale, convert the working copy first to grayscale (Color→Color to Grayscale), then to 16 bits/pixel (Color→Change Image Depth). Invert the grayscale values if necessary (Ctrl-V) so the bands are black on a light background. ALWAYS use an original 16-bit grayscale image if possible. This is the only format suitable for image analysis.
It is good practice to create a histogram of your image (Color→Histogram) before starting to determine the quality of your image. A good image will have a histogram that resembles a forest of lines. If there are only a few widely-spaced lines, the image might not be good enough to analyze. An image grabbed from a PDF, a word processor, or Powerpoint might look fine to the eye but be useless in forensic analysis.
These types of images are not suitable for analysis:
Color images
8 bit/pixel colormapped images
Images containing artifacts such as text obscuring the bands
Low contrast or smeared images
Select Color→Grayscale map and set the sliders to the widest range. If this causes the image to be too dark or too light, it means the image does not have sufficient dynamic range for analysis.
Click on Measure→Detect Image Manipulation. The first two tests are the principal tests. The others are extra tests that may be useful to confirm or refute any conclusions from the initial test.
In some cases it might be necessary to enhance the contrast of the image before it can be properly analyzed. Be sure to document what was done, and work only on the copy.
After analysis, hit Ctrl-R or click the Undo button to restore the un-annotated image.
Specific instructions
Click on Measure→Detect Image Manipulation. The options are as follows.
Basic Histogram analysis
One image is used at a time in histogram analysis. The software detects the bands automatically using a segmentation algorithm and compares each band to all the other bands in the image.
Basic histogram analysis creates a histogram for each band. Then it performs a linear regression between each pair of histograms. The advantage of this method is that rotation, flipping, and warping of the band do not affect the result. The disadvantage is that if the miscreant changes the brightness or contrast, the similarity is not detected, and the more advanced option (below) is needed.
The results are shown in a table showing the F value and correlation coefficient (r-squared) of each comparison. An r-squared value of 0.98 or higher could be evidence of a copy or copy/flip operation. A value of 1.0 indicates an exact copy or copy/flip. If every line in the table is above 0.99, it probably means the image was converted incorrectly.
Band detection setting
Automatic: finds the bands automatically. Their size will vary.
Automatic fixed size: finds the bands automatically but analyzes the rectangular region specified under Width and Height. This is the default.
Manual: Allows manual selection of bands (not yet implemented).
Labels
Labels are shown in red and have no effect on the analysis. The labels can be removed by pressing Ctrl-R or clicking the Undo button. You can select how much labeling you want.
Signif. r2 threshold
The table shows pairs of bands with a correlation coefficient greater than the specified threshold. The user should practice changing this setting because the correct threshold will vary with the quality and size of the image. In most cases, an r2 above 0.95 indicates something worthy of additional inspection, while a value of 0.98 indicates a close match. If a band was copied exactly (even if flipped) the r2 is 1.0, meaning a perfect match.
Pattern match weight and pattern mismatch weight
If the bands are not identified correctly, adjust these two parameters and repeat the analysis until each band is in a separate red box. If the image is too dark, or if the bands are not well separated, two or more bands may be run together. If nothing else works, draw a white line between them ON THE COPY to separate them.
Filename for histograms
Check this box to save the histograms for each band in the specified file in text format. These can be plotted in a graphing program to document the similarity or lack of similarity of any two bands.
Smooth histogram
The histogram will be smoothed by a Gaussian 21-point smoothing function before analysis. This may increase the accuracy in images where the bands are too small to produce a good histogram.
Histogram analysis with scaling detection
This is similar to an ordinary histogram analysis except that the histograms are analyzed in a different way to detect a contrast and brightness change in addition to a copy or copy/flip. This takes a fair amount of memory and calculation, so it may take several seconds to analyze an image. Miscreants often lighten or darken a band when copying it to conceal their actions. This option calculates the pixel value scale factor and offset that they may have used. The table is also slightly different than the table in the basic option in that it prints the contrast and brightness factor that was applied to the image.
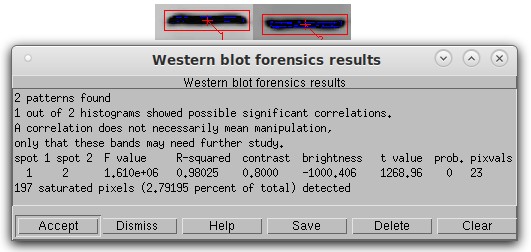
Histogram analysis with scaling detection. In this simulated Western blot, Band 2 is a copy of band 1 that was flipped vertically, reduced in contrast by 20%, and had a pixel value of 1000 subtracted from it. Despite the presence of 197 saturated pixels (blue), the algorithm found the contrast, brightness, and flipping manipulation with no problem with an r-squared of 0.98 and a p-value of 0.0. The last column shows the number of discrete pixel values actually used in the calculation as a control check of the algorithm.
Saturated (pure black or pure white) pixels should always be minimized in a scientific image. They can have a deleterious effect on any image analysis, so after the calculation the program tells you the number and percentage of saturated pixels. It also shows all pixel values of 0 as blue and all pixels saturated at white as red. These colors are superimposed on the display and do not affect the actual image.
Filename for output histograms
If this option is selected, all the intermediate histograms will be written to the specified text file for inspection. Be warned, this file can be very large.
Smooth histogram
The histogram will be smoothed by a Gaussian 21-point smoothing function before analysis. This may increase the hit rate in images where the bands are too small to produce a good histogram. A minimum of 100 different gray values (About→About the image) is recommended.
Find differences
Two images are needed for this option. It highlights differences between two images (Suspect image and Control image). If they are identical, the output image is solid purple. If not, the pixels are color-coded to show where they are different. The two images must be perfectly aligned before this option is used. (Edit→Shift).
Butterfly plot
Two images, each containing a single band, are needed for this option. A butterfly plot creates a diagram showing the degree of similarity between two images. If they are identical, the graph is a solid diagonal line. Some image analysts assert that the correlation coefficient of this line is related to the degree of similarity. However, this is debatable, and the butterfly plot is extremely sensitive to small differences in position of the bands within the two images, as shown here.
Since these plots tend to be rather faint, you can make the individual pixels bigger by checking Heavy Pixels under Appearance.
Horizontal profile difference plot
Two images, each containing a single band, are needed for this option. Unlike the butterfly plot, perfect alignment is not essential. The horizontal profile plot uses densitometry to trace the two images from left to right. If the bands are identical, the traces will be identical. Checking the Toggle Button next to Density Trace will save the densitometry traces in text format under the filename you specify. Any graph drawing program can read these files and plot them as desired.
To create the two images from a larger image, click the New button at left, click Fixed Size, and set the x and y size of the image to be created. Click Yes under Use Same Filename if you want the new image to retain the same filename as the larger image. Then click Accept and click on the upper left point of the desired image.
When plotted in graph-drawing software, a flat horizontal line produced by subtracting the two densitometry traces indicates a perfect match that implies copying. More information is available by plotting the un-subtracted traces. If one band is a flipped copy of the other, the horizontal line will be a mirror image of the original band. If a miscreant made contrast or brightness adjustments to hide the copying, the curves are simply shifted in position with little effect on their overall shape. This makes it easy to identify what transformations have been applied to the bands.
More calculations and statistics on these datasets can be done in a spreadsheet program such as xdata.
Vertical profile difference plot
Same as horizontal, except scans vertically (still under development).
Zebra plot
This option is under development. Applies a black and white colormap or a pseudocolor map (sometimes called a density map) to the image to detect any edge artifacts produced by image manipulation. This method uses only a single image at a time.
Until development of this option is complete, you can perform the same operation by following these steps.
Color→Change image depth→Convert to 8 bits/pixel
Color→Colormap/false color - selects the colormap to be applied to the image.
Select Colormap: provides a variety of colormaps which can be rotated. Select gray scale/false color first. Over 1000 colormaps are available.
Color→Convert Image to Color: changes the 8-bit grayscale image to 8-bit indexed color. Check the image under About→About the image to verify that the Color type is now Indexed color.
Then go back to Colormap options, select colormap, and pick Zebra or Other. By dragging the slider in the box titled Enter Colormap Number, you can change the colormap as needed to show any edge artifacts.
Another way to find edge artifacts is to filter the image using a 3×3 Laplace filter (Edit→Filter→Filter type=Laplace Edge Enhancement). Set the Amount of Filtering to 100% and leave the other settings at their default values. An unmanipulated image will become slightly fuzzy when this filter is applied. A manipulated image will show edge artifacts where manipulation may have occurred.
Note that miscreants will not necessarily copy a rectangular area, so these edges will not necessarily show up as straight lines. Many software programs including Imal allow selection and copy/paste of arbitrary-shaped areas.
Also note that once the image has been converted to 8 bits/pixel, the 16-bit (4.8-log) dynamic range of the original image is permanently lost. Another copy of the image must be reloaded before other tests can be done.
Smoothness map
Smoothness detection is valuable because miscreants may artificially smoothen parts of the image to conceal edge artifacts created by manipulation. These are very difficult to see in the original grayscale image. Thus, if a highlighted area shows up on the smoothness map, it could indicate that some manipulation may have occurred in that region.
Areas that are smooth are highlighted in red, while areas that are sharper are highlighted in blue. This highlighting does not affect the image and can be cleared by hitting Ctrl-R.
The Range parameter can be used to adjust how much smoothness is detected. A higher range value will find larger areas of smoothness.
Densitometry
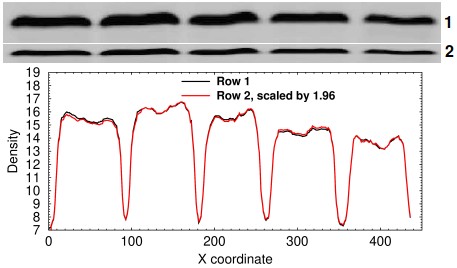
Densitometry analysis of a resized row. In this simulated example, row 2 was made by copying row 1 and shrinking it vertically by 50%.
It sometimes happens that a miscreant simply copies a band or an entire lane and resizes it vertically or horizontally. This can be tested by doing horizontal or vertical strip densitometry, plotting the traces, multiplying the resized band by the ratio of the sum of the densitometry signals, and performing a linear regression. In the example at right, the two lanes are strongly suggested to be identical by a coefficient of correlation of 0.999194 and by the fact that the two curves are easily superimposed.
Resizing an entire row might sound like a dumb thing to do, but I have seen it happen. If the individual also flips the row, it can easily be missed. This sort of thing is hard to do by accident and suggests an attempt at deception.
Future additions
Future versions of Imal will have additional functions that detect similarities in shape. These are generally of more limited value in studying Western blots because many suspicious images are only available as copies of images obtained from PDF files. Unfortunately, most commercial software converts every image either to 24-bit RGB color or 8-bit grayscale when it produces a PDF file. The image might appear identical to the original by eye, but in fact its dynamic range has been greatly reduced. The down-conversion also creates artifacts, which can make shape comparisons difficult and can also produce false positives. However, if shape detection can be adapted to correct for changes in size and aspect ratio, it can be a powerful tool.
sep 02 2022, 3:34 pm
