 or years it was dogma that the observed sex differences in Alzheimer's disease
(AD) were solely due to women's longer lifespan. Men died at an earlier age from
other causes, mainly work- and stress-related, and it was thought that this could
explain why two-thirds of AD patients were women. But after women entered the
workforce and were subjected to more workplace stress, the ratio barely budged.
Now research using a new technique shows us why: women really are more susceptible
to AD. It's an important clue that points directly to the
immune system.
or years it was dogma that the observed sex differences in Alzheimer's disease
(AD) were solely due to women's longer lifespan. Men died at an earlier age from
other causes, mainly work- and stress-related, and it was thought that this could
explain why two-thirds of AD patients were women. But after women entered the
workforce and were subjected to more workplace stress, the ratio barely budged.
Now research using a new technique shows us why: women really are more susceptible
to AD. It's an important clue that points directly to the
immune system.
RNA-seq
The new technique is called RNA-seq, also known as transcriptomics, where you take the RNA, reverse-transcribe it into DNA, add a special linker, and then sequence the DNA using a technique called next-generation sequencing to find out what proteins are being expressed in your sample. You then look for clusters of genes that have related functions.
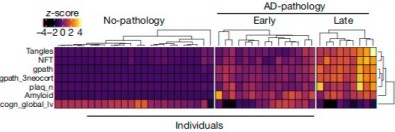
RNA-seq results (from Mathys et al. [2])
This new technique has become something of a fad, just as DNA microarrays were two decades ago, but it has quite a few limitations. For AD, the main limitation is that RNA is rapidly degraded after death, so only freshly collected brain samples are usable, and even those may be questionable because it's well known that some mRNAs are degraded faster than others. To avoid this problem, you can use nuclear RNA, which is said to be less rapidly degraded, but this adds a new variable to the equation because RNA in the nucleus might be different from RNA in the cytosol.
If the technique works, it will tell you you've overlooked some important biochemical pathway. If it doesn't, it tells you what you already know.
As you might expect, sequencing the tens of thousands of RNA molecules generates a lot of data, so elaborate computer processing by bioinformatics suites like Bioconductor is needed to make sense of it. People have to use innovative data visualization techniques just to make sense of the results. It's a monumental task which requires monumental amounts of money. Since it's technique-driven, typically the first study gets into Science or Nature and subsequent studies soon become unpublishable unless you improve the technique.
One paper in Neuron [2] tried a clever approach: use transgenic mice in which the native mouse apolipoprotein E gene has been replaced by one of the three human apoE genes, and then validate their results by comparing them with RNA-seq results done by somebody else. Here are their findings:
- The three different apoE genotypes changed the expression of 1813 genes, mostly in genes like Serpina3n, which was already known to be associated with AD.
- Age changed the expression of 10,244 genes.
- Sex changed the expression of 78 genes, mostly in proteins involved in lipid metabolism.
Sex differences
Can RNA-seq tell us anything more specific? Yes indeedy. The main paper on this topic is from Mathys et al.[1] who sequenced 80,660 single-cell transcriptomes from 48 patients and put their results in the ROSMAP database for other scientists to use. This was an enormous amount of work: each sample was obtained from a single cell (which they isolated using a fluorescence-activated cell sorter). They reported changes by cell type, disease severity, and sex. Here are their findings:
- 96% of the changes in AD were found only in one specific type of cell (astrocytes, microglia, excitatory neurons, etc.). This means every kind of cell in the brain is affected in a different way in AD. Most of the changes in excitatory and inhibitory neurons were downward, while changes in other cells were mostly upward. ApoE was upregulated in microglia but downregulated in astrocytes. These are big findings, but to researchers it also means that to do RNA-seq they don't just need a hideously expensive Next-Generation Sequencing setup but also a very expensive cell sorter.
- Changes in RNA expression start before severe pathological features of AD.
- As feared, many of the changes were correlated with post-mortem interval and age of death, but most of these changes were different from the ones that correlated with AD.
- Genes that were good markers for AD pathology (such as neurofibrillary tangles, beta-amyloid, and plaques) were more highly expressed in females than in males. Both sexes had these changes in neurons, but females had a much bigger change than males. Males also had changes in oligodendrocytes, which produce the myelin sheath that covers axons, but females didn't. The myelin sheath makes up the white matter in the brain.
Inflammation
The oligodendrocyte finding is important: it hints at even stronger connections between AD and the immune system than previously thought, because oligodendrocytes are the target in multiple sclerosis, which is an inflammatory disease. The sex differences also give us an important clue. They confirm the idea that females are more susceptible to AD than males, though why this should be is not yet clear.
To laymen, this may just seem like bad news, but finding a connection between two terrible diseases like MS and AD qualifies as a minor breakthrough.
Already a bunch of papers are reporting that HLA (human leucocyte antigen) proteins may also be involved in AD. These proteins are MHC (major histocompatibility complex) class I or II proteins and are part of the adaptive immune system. A polymorphism in HLA-DRB1 was found by genome-wide association studies to be associated with AD, and other HLAs appear to be substrates for presenilin and α-secretase. No one knows what these findings mean yet. Most of the articles are behind heavy paywalls demanding $39.95 or so to read them, so researchers tend to skip over them.
New definitions
But maybe we ought to pay up and read them. An example is a paper in the Journal of Alzheimer's Disease almost four years ago. Many of us don't read this journal because most libraries don't have it, so we have to wait a week or so for Interlibrary Loan to find it for us. Those guys were all in lockdown because of some kind of virus or something, so it was only by chance that I saw it.
It tells us something that's been incubating for a long time: sporadic (late-onset) AD is shaping up to be a completely different disorder than familial (early-onset) AD. Most of the mutations in early-onset AD are not in APP, but in presenilin 1. APP, which is also called amyloid precursor protein or sometimes AβPP, is cleaved to produce beta-amyloid. Presenilin 1 is a protease that has over 90 different substrates, which means it's involved in a fiendishly complicated bunch of pathways in the brain, any or all of which could contribute to AD.
One of these substrates is APP. Presenilin cuts APP to make beta-amyloid, but contrary to what you might think the mutations don't make it more active. For a while people thought that the mutations increased the ratio of bad beta-amyloid (Aβ42) to less-bad beta-amyloid (Aβ40), but that theory never made sense and it was eventually shown not to be generally true. Drugs that block presenilin can actually make AD worse—the opposite of what you'd expect. Writing in JAD, A. E. Roher finds many other incompatibilities between presenilin-familial AD (PSEN-FAD) and sporadic AD (SAD). He writes:[3]
Amyloid deposits and NFT [neurofibrillary tangles], present in many other neurodegenerative diseases, do not necessarily represent the primary and only causative factors in PSEN-FAD and SAD pathogenesis. An alternative explanation that can be seriously contemplated is that amyloidosis represents a common denominator in the phenotypic expression of PSEN-FAD and SAD. This line of argument may lead to the tantalizing possibility that in these two types of neurodegeneration Aβ may have a still undefined rescue function, rather than having a solely pathogenic role.
Many patients with AD, Roher says, have no amyloid deposits and no detectable soluble amyloid. Many others have ‘mixed’ pathologies. He calls for redefining AD into three new diseases: SAD, AβPP dementias, and presenilin dementias.
Not everyone, least of all influential researchers like D. J. Selkoe, who still talks about reinvigorated support for the amyloid hypothesis,[4] is convinced of the need for that yet, but it looks more and more like beta-amyloid may not be a direct cause of AD but a response to inflammation, infection, or injury.
Why is getting the names right so important? To paraphrase Confucius, if you don't define your disease right, you won't be able to cure it.
An experiment we tried years ago drove that point home for me: we put catalase, an innocuous protein that breaks down hydrogen peroxide, on our cells at the same concentration that people use for beta-amyloid. Why we tried that is a bit fuzzy now, but it killed the cells dead. Maybe there's also a lesson about testing the specificity of your toxic proteins in there somewhere.
1. Zhao N, Ren Y, Yamazaki Y, Qiao W, Li F, Felton LM, Mahmoudiandehkordi S, Kueider-Paisley A, Sonoustoun B, Arnold M, Shue F, Zheng J, Attrebi ON, Martens YA, Li Z, Bastea L, Meneses AD, Chen K, Thompson JW, St John-Williams L, Tachibana M, Aikawa T, Oue H, Job L, Yamazaki A, Liu CC, Storz P, Asmann YW, Ertekin-Taner N, Kanekiyo T, Kaddurah-Daouk R, Bu G (2020). Alzheimer's Risk Factors Age, APOE Genotype, and Sex Drive Distinct Molecular Pathways. Neuron 106(5), 727–742.e6. doi: 10.1016/j.neuron.2020.02.034. PMID: 32199103; PMCID: PMC7388065.
2. Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X, Martorell AJ, Ransohoff RM, Hafler BP, Bennett DA, Kellis M, Tsai LH. Single-cell transcriptomic analysis of Alzheimer's disease (2019). Nature 570(7761), 332–337. doi: 10.1038/s41586-019-1195-2. PMID: 31042697; PMCID: PMC6865822.
3. Roher AE, Maarouf CL, Kokjohn TA (2016). Familial Presenilin Mutations and Sporadic Alzheimer's Disease Pathology: Is the Assumption of Biochemical Equivalence Justified? J Alzheimers Dis. 50(3), 645–658. doi: 10.3233/JAD-150757. PMID: 26757189.
4. Aisen PS, Cummings J, Doody R, Kramer L, Salloway S, Selkoe DJ, Sims J, Sperling RA, Vellas B (2020). The Future of Anti-Amyloid Trials. J Prev Alzheimers Dis. 7(3), 146–151. doi: 10.14283/jpad.2020.24. PMID: 32463066.
nov 01 2020, 7:05 am
