 an Li-Meng's second paper, in which she and her colleagues promised to show
that RaTG13 (the virus said to be the closest relative of SARS-CoV-2) is fake,
came out last week. Compared to the first paper[1], this one has
made almost no impact. Why? Because it's a mess.
an Li-Meng's second paper, in which she and her colleagues promised to show
that RaTG13 (the virus said to be the closest relative of SARS-CoV-2) is fake,
came out last week. Compared to the first paper[1], this one has
made almost no impact. Why? Because it's a mess.
The second paper[2] is 33 pages long. It has 123 references (though some are simply tweets and web links) and it's very frustrating to read because the main points are so obscured, so let's break it down into small chunks.
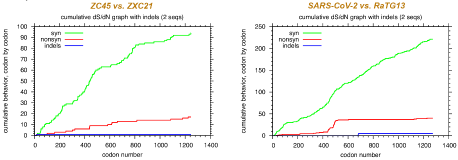
Fig. 1 Mutation plots of SARS-CoV-2 vs RaTG13 from Yan et al. [2]
Their primary evidence is the ratio between synonymous and non-synonymous mutations in the spike protein. In this plot (Fig. 1), two different DNA (or RNA) sequences are aligned and the cumulative number of synonymous and non-synonymous mutations is plotted. On the left is ZC45 vs ZXC21. On the right is SARS-CoV-2 vs RaTG13. The y-axis scale is different in the two graphs, but what they show is that ZC45 and ZXC21 differ synonymously at 94 sites and non-synonymously at 17 positions (ratio=5.5 to 1), while SARS-CoV-2 and RaTG13 differ synonymously at 221 positions and non-synonymously at 41 positions, so the ratio is 5.4 to 1—almost the same.
A non-synonymous mutation simply means it codes for a different amino acid. A synonymous mutation means the DNA is different, but the amino acid is the same. It's what most people call a conservative mutation. The red curve in the second graph jumps up at around codon 500, which is just before the S1 and S2 regions are joined by the infamous furin cleavage site, and then remains more or less flat, so the authors calculate that the ratio in S2 is 44.0 to 1. This, they say, is suspicious.

Fig. 2 Mismatches and insertion in SARS-CoV-2 vs RaTG13
To find out what in the world they're talking about, I had to download and analyze the sequences myself. Here is what I found.
I'm a protein guy, so it's easier for me to look at the peptide sequence. To analyze it, you have to examine the original sequences because there are many new sequences in the database that have been modified to excise the S1/S2 cleavage site. These are called pseudoviruses and they've been engineered to make the virus safe to work with.
In an identity plot (Fig. 2), a mismatch shows up as a broken line and an insert or deletion (called an indel) shows up as a shifted line. There's only one insertion, shown in the lower panel, which occurs at the furin cleavage site around codon 680. The variable region (upper panel) occurs much earlier, around codon 500. Walls et al.[3] describe this mismatch region clearly in their Cell paper comparing SARS-CoV-2 to SARS-CoV. What is this domain? It is the receptor-binding motif in S1.
The mismatches are shown in Fig. 3. It is the region you'd expect a sinister person to mutate if they wanted to make the virus more hazardous to humans. But it's also the region you'd expect a virus to mutate as it evolves in whatever species it evolved in to increase its pathogenicity. The red curve in Fig. 1 remains flat because it is in S2, which is not involved in receptor binding, so there is less evolutionary pressure on S2. So the graph in Fig. 1 is not probative in any way. It merely means that the protein became adapted in humans or human cells. Discovering how that happened will be the key to uncovering its origin.
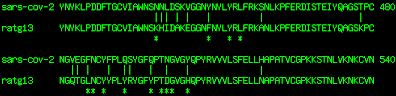
Fig. 3 Variable region in SARS-CoV-2 RBM. Amino acid residues essential for binding to the human ACE2 receptor are shown with stars.
Other evidence
Okay, so what other evidence do they have? Some of it is circumstantial: they ask why the famous case of the Mojiang miners didn't get more attention. They point out, correctly, that the intermediate host (if there is one) has not yet been identified. And they complain that the virus samples are contaminated with DNA from other species.
At best, these claims show that the work done by the Wuhan Institute of Virology was not up to Western standards. If you or I were doing their job, with the benefit of hindsight, we might have collected better samples. We certainly would have paid more attention to the fate of the miners. Perhaps the Bat Lady did some sloppy work, or perhaps they were in a hurry. But diagnosing diseases and discovering viruses is not as easy as people think.
It also means little, as some people are claiming, that the CDC and WIV don't have samples of the live virus. Nowadays, a virus can be discovered and characterized without ever isolating it. That doesn't mean that it's not real.
In the rest of the paper, they accuse the WIV lab of inventing RaBtCoV/4991 (the original forgotten partial sequence of RaTG13) as well as two pangolin viruses (MP789 and P4L), and a new virus sequence RmYN02, which is said to be 93.3% identical to SARS-CoV-2.
Comment
The main mystery of the four amino acid insert which creates a furin cleavage site remains unexplained. The remaining evidence, presented in this paper, that RaTG13 is fake, is unconvincing. The paper is tendentious and is excessively political in its approach. Although we may be sympathetic to the author's ideas, this paper only makes it harder for scientists to examine the evidence. While we can blame the PRC government for their handling of the virus, you cannot prove an accusation by making more accusations. An old saying comes to mind: if you strike at the king, you must kill him.
1. Yan LM, Kang S, Guan J, Hu S (2020). Unusual Features of the SARS-CoV-2 Genome Suggesting Sophisticated Laboratory Modification Rather Than Natural Evolution and Delineation of Its Probable Synthetic Route. https://zenodo.org/record/4028830
2. Yan LM, Kang S, Guan J, Hu S (2020). SARS-CoV-2 Is an Unrestricted Bioweapon: A Truth Revealed through Uncovering a Large-Scale, Organized Scientific Fraud https://zenodo.org/record/4073131 The 2nd Yan Report.pdf doi 10.5281/zenodo.4073131
3. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020 Apr 16;181(2):281-292.e6. doi: 10.1016/j.cell.2020.02.058. Mar 9. PMID: 32155444; PMCID: PMC7102599.
oct 13 2020, 6:46 am
